RZSS WildGenes New Years resolution – find out what a wild animal really eats
04/02/2018 in RZSS

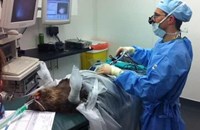
Above: RZSS WildGenes are using faecal samples from Sumatran tiger Jambi at Edinburgh Zoo to test their new analysis methods for establishing the diets of wild animals.
With the winter festivities now a distant memory and the seasonal goodies long since devoured, the first few months of the year are occupied with new year’s resolutions. Along with joining a gym and/or saving money, many resolve to eat a healthier diet. Whilst animals are not conscious of their waist size, maintaining a healthy and appropriate diet is still important for species to survive and thrive in their environment. With an increasing number of animals under threat, a thorough understanding their ecology can ensure that conservation management plans are suitably tailored. Knowing precisely what and where a species eats can be critical to managing populations in captivity as well as helping to conserve animals in their wild habitat, especially where they live near humans and can be accused of predating livestock and farm crops.
Fortunately, we don’t need to rely upon animals keeping a ‘food diary’ to understand their diet, instead we can use cutting edge DNA technology to understand what items an individual or species has eaten. Insights into diet can be gained by examining fragments of undigested items that get passed as faecal material, such as seeds, bits of bone, and hair/fur or feathers. Although this can give an indication to the types of food that are consumed, many items can be difficult to identify even with specialist knowledge and equipment.
Metabarcoding is a form of analysis which looks at the DNA present in a faecal sample, including traces from food items which an animal has eaten. The fragment of DNA chosen, is one that is good at distinguishing between different species, usually a portion of the mitochondrial or chloroplast genome. It is amplified (using a PCR - polymerase chain reaction) and then sequenced using next generation sequencing technology. The DNA sequences are sorted, filtered, and compared to a database which contains all known possible species sequences. When a sequence returns a positive match against the database we can then identify which species were consumed by the animal and we are using this technology to analyse the diet from a range of species.

Above: Jen Kaden (RZSS) and Dr Linda Neaves (RBGE) in the RZSS WildGenes laboratory, extracting DNA from faecal samples collected at Edinburgh Zoo.
This technique is being applied to take a closer look at the diets of the giant panda and other herbivores, both in captivity and in the wild. Whilst most can say that pandas eat bamboo, there are over 60 species of bamboo available to pandas in their native range and conservation managers want to know which the animals prefer. In collaboration with Dr Linda Neaves and the Royal Botanic Garden Edinburgh we are testing new methods that will be able to help us distinguish the different species of bamboos because previous analysis has found it difficult to accurately tell them apart. If successful this information will be vital in helping us to understand the relationship between pandas and bamboo, not to mention any other food sources that may be part of their diet.
Understanding the true composition of wild animal diets has traditionally been difficult to research. For example, many species that are elusive or predate at night are difficult to observe. When communities that live near to areas inhabited by wild carnivores also own pets or livestock, the animals could seem easy prey for a carnivore compared to hunting wild prey items; a bit like targeting a take-away MacDonalds instead of chasing around a supermarket for salad! When wild animals do take livestock, or when loss is blamed on wild animals, it can result in a human-wildlife conflict that negatively affects both the community and the accused species. Analysing faecal samples in the laboratory should make this process significantly easier. Through determining what a carnivore has eaten, we can then inform strategies for minimising situations of conflict and help optimise the structure of protected areas. With this aim in mind, we are developing our methods using faecal samples collected from Jambi, RZSS Edinburgh Zoos resident tiger. We can test the accuracy of our results against the dietary information provided by his keepers and work out how best to use this ‘tool’ for wild animals samples.
Although wild animals do not have a list of ‘new year’s resolutions’ to try and keep, knowledge of their diet composition is likely to help benefit their conservation management. No matter how hard we try, it is always difficult to substitute a carrot stick in place of a tasty chocolate bar. Whilst it is easy to convince ourselves that we have stuck to our new year’s resolution, using a DNA diet assessment to reveal what we have really eaten might just explain why we can’t shed those few festive inches!
Until next time
Dr Gill Murray-Dickson (RZSS Research Scientist) and Jen Kaden (RZSS Senior Lab Technician)
Further reading:
RGBE: Using DNA to investigate giant panda diet
RGBE: Panda diets and conservation
Support our work
By becoming an RZSS Member you can help support our conservation work around the globe and enjoy unlimited access to Edinburgh Zoo and the Highland Wildlife Park for a whole year.



























Follow EZ